I must have been really busy these last few weeks to have gone so long without posting about this paper. For anyone interested in the microbiome this is a hugely important paper to read (link). If you want a more comprehensive summary, you can find a few in the popular press.
At the risk of rambling on and on, I want to talk about a few things from this paper that really caught my eye. Let's start with Figure 1a.
Figure 1a: Personalization, niche association, and reference genome coverage in strain-level metagenomic profiles. a, Mean phylogenetic divergences17 between strains of species with sufficient coverage at each targeted body site (minimum 2 strain pairs)
The statistic being plotted is the "mean distance of strains," meaning that they computed the exact genome sequence of each of the strains of all of the dominant organisms in every sample(!) and then calculated how different the strains within each species were between different samples. That process is (in my opinion) a difficult task to pull off well, and I find myself in the position once again of being very much in awe of the Huttenhower group for their skill and hard work. Ok, now how about the biology? This figure tells us that people harbor strains of microbes in their microbiome that are distinct from other people's strains, and that those strains stick around over time to some degree. Not only that, but the degree to which those strains stick around varies by body site, with the stool (and gut) likely having the most persistent set of strains.
Ok, so now we've covered the fact that people have different strains of the same microbial species in their microbiomes, so let's go into a bit more depth with more of figure 1.
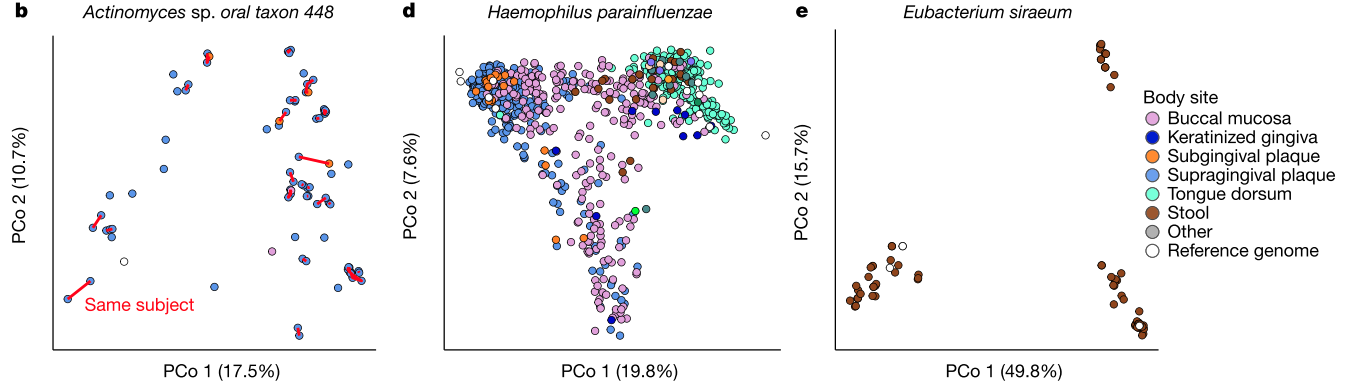
Figure 1, continued. b, Individuals tended to retain personalized strains, as visualized by a principal coordinates analysis (PCoA) plot for Actinomyces sp. oral taxon 448, in which lines connect samples from the same individual. d, PCoA showing niche association of Haemophilus parainfluenzae, showing subspecies specialization to three different body sites. e, PCoA for Eubacterium siraeum.
Let's break this out:
- Figure 1b: The exact strain of (one of the) bacteria in your dental plaque sticks around from day to day, even though people are (presumably) brushing their teeth!
- Figure 1d: A bacterial species that is found all over the body, H. parainfluenzae, is genetically distinct (to some degree) by body site. Most intriguingly, it is only partially distinct by body site, which raises all kinds of questions about its evolutionary history.
- Figure 1e: An organism that we call a single species (E. siraeum) seems to form three completely distinct genetic groupings. Note that the horizontal axis accounts for ~50% of the total genetic variation. That's huge. Is it a single species? Is it three? What is a "species"? Does it matter?
Reflections:
I don't want to try to sum up this paper with a single take-home message. I think this is a paper to read and reread and think about. However there are a few aspects of the methods used that I want to point out for those who may not think about this type of analysis very often. The first is that the authors defined a single strain for each sample (using StrainPhlAn). Do we think that there is only one strain of each species present at a single time? How could we test that hypothesis or even deal with a sample containing multiple, closely related strains? The next is that their most in-depth characterization of strain differences hinged on comparing the samples to known reference genomes. What about the variation that has never been captured in a reference genome? How would we even approach that data?
Lastly I'll say that I really think this work is important because I think that strain level variability in the microbiome is a crucial factor in human health and disease. This paper provides strong evidence that strain level variation is extensive, and the authors have provided powerful tools for characterizing that variation. The next question is, how do we apply this type of data in a way that uncovers the biological mechanisms underlying human health? In other words, how do we use microbiome profiling to generate some experimentally testable hypotheses? It seems so clear to me that this avenue of research is going to uncover important biological mechanisms of the human microbiome, but I can also see that we're going to need some creative, collaborative problem solving in order to realize this system's full potential.
UPDATE:
On October 30, 2017, the Pollard Lab posted a preprint in which they specifically tackle the challenge of analyzing multiple strains per species in a given sample. You can read the preprint here. There's a lot of detail there that really deserves its own blog post, so stay tuned.